Post provided by Roberta L. C. Dayrell
The knowledge of seed morphology is an essential resource for practitioners and scientists across diverse disciplines such as botany, agriculture, restoration, conservation, and archaeology. Morphological attributes can inform studies on topics such as seed dispersal, predation, longevity, and germination. This knowledge also has practical applications, including seed identification and assessments of quality and ripeness. But extracting meaningful information from these structures can be trickier than it seems!
Challenges in Seed Morphological Analysis
That’s what we found when we started looking into objective and meaningful ways to describe the morphological characteristics of seeds from a variety of species and plant families. Manual measurements of seed size are time-consuming and provide limited attributes to communicate seed morphology. On the other hand, visually categorising seeds based on colour and shape, for example, is a relatively quicker process, but the results can be subjective, especially when the differences between seeds are subtle.
Image analysis methods offer a viable alternative to tackle the problem of ‘seed secrecy’ and obtain fast and objective characterisation of biological objects. However, there are challenges in automating the process of trait extraction. Separating seeds from the background is difficult when working with objects of varying physical attributes. Morphological differences in seeds cause optimal parameters to vary, requiring users to fine-tune the settings for each species. Extracting meaningful information is also a challenge, as most software only provide a few standard measurements or require coding knowledge.
Optimising trait extraction
To address these challenges, we have developed a method which consists of a simple image acquisition and processing protocol coupled with an open-source software called Traitor. The workflow for trait extraction involves scanning seeds against a high-contrast background with a flatbed scanner, correcting image colours, followed by a fully automated image analysis with the software.
Digital images are made up of pixels. Traitor uses k-means clustering, a type of unsupervised machine learning algorithm, to group similar pixels together into two groups: seeds and background. This is why scanning seeds against a high-contrast background is crucial to ensure the algorithm groups all those background pixels together, effectively separating them from the seeds! After this separation, seeds are aligned, and several morphological traits are extracted. Thus, when given an image of seeds with a high-contrast background, Traitor will do what a traitor does best: carry out automatic trait extractions and ‘expose’ seeds by giving away their valuable information.
Traitor was built on Python, but the good news is no prior knowledge is required from users! Once the software is installed, entering some basic commands in the computer’s prompt or terminal is all it takes to extract the traits. We have carefully written instructions for installation and use, so check those out here
Interpretable output
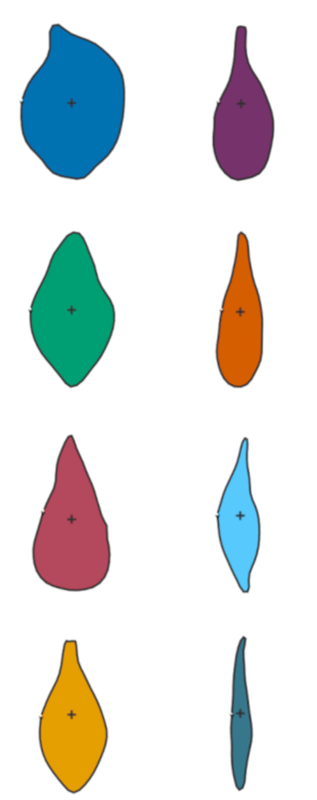
Our paper brings examples of how Traitor’s output can be used to improve the communication of seed morphology and advance on new identification tools. We used Traitor’soutput to obtain representative colours for seeds in the Rosaceae family and to build objective categories for seed shape for Carex species. We also showed how we can use extracted traits to answer evolutionary questions, by assessing the degree to which colour measurements are correlated with the evolutionary relationships among species within the Asteraceae family.
Moving forward
We hope that this method will help enhance the communication and understanding of seed morphology while paving the way for the development of innovative identification tools. Moreover, it offers exciting opportunities for exploring the relevance of morphological traits in the fields of ecology and evolution.