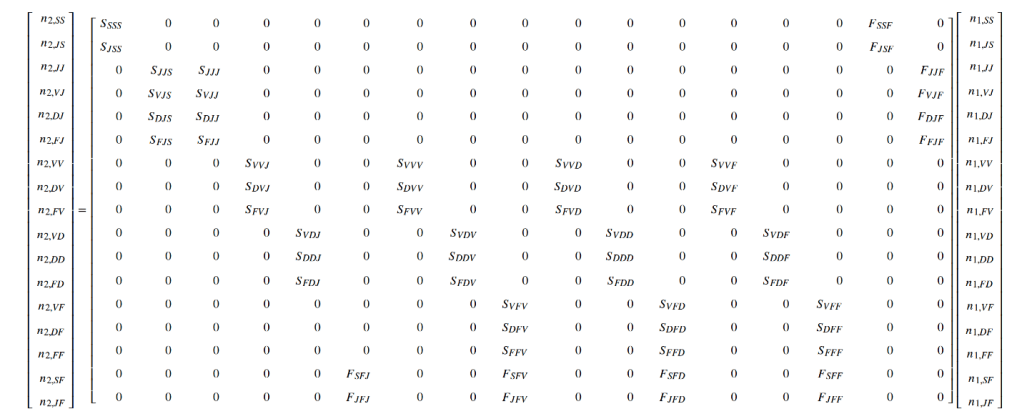
How to write clean code
Alessandro Filazzola & Christopher Lortie tell us about their new article ‘A call for clean code to effectively communicate science‘, which provides a series of recommendations and a suite of tools that can be used to help support scientists to produce cleaner code. Can you clearly understand the code that you have written? What about if you gave it to a colleague? Or a reviewer? … Continue reading How to write clean code