Post provided by Rich Shefferson
Matrix projection modeling is a mainstay of population ecology. Ecologists working in natural area management and conservation, as well as in theoretical and academic realms such as the study of life history evolution, develop and use these models routinely. Matrix projection models (MPMs) have advanced dramatically in complexity over the years, originating from age-based and stage-based matrix models parameterized directly from the data, to complex matrices developed from statistical models of vital rates such as integral projection models (IPMs) and age-by-stage models. We consider IPMs to be a class of function-based MPM, while age-by-stage MPMs may be raw or function-based, but are typically the latter due to a better ability to handle smaller dataset. The rapid development of these methods can leave many feeling bewildered if they need to use these methods but lack sufficient understanding of scientific programming and of the background theory to analyze them properly.
lefko3 R Package
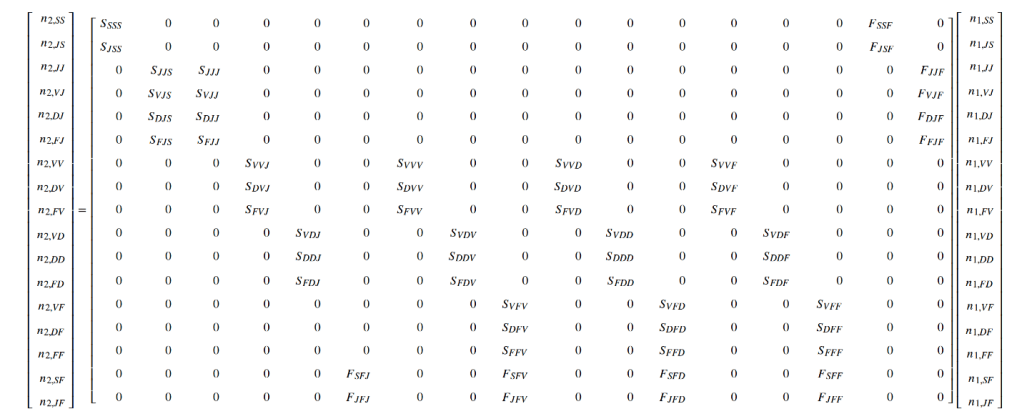
R package lefko3 is an attempt to make this world more accessible by providing powerful tools that automate much of the process of creating and analyzing MPMs. It is particularly focused on historical MPMs (hMPMs), which are more difficult to parameterize properly, although it also comes with tools to create most other kinds of MPMs. The historical MPM is a matrix population model that breaks one of the key assumptions of matrix projection modeling – that the state of an organism in the next time is determined solely by its current state. This assumption contradicts several decades of evidence showing that events in the life of an organism, including memory, environmental conditions and life history strategy, have large effects in the long-term, sometimes even affecting future generations. Historical MPMs assume that state in the next time is determined by current state as well as the state of the organism in the previous time step. This gives hMPMs the ability to incorporate life history relationships such as costs of growth, which are not possible to model in normal “ahistorical” MPMs (ahMPMs). It also makes hMPMs much larger and much more sparse than normal “ahistorical” MPMs. For example, an ahistorical MPM with 10 life history stages will be 10 rows x 10 columns = 100 elements large, with potentially all values non-zero. In contrast, the associated hMPM will be 100 rows x 100 columns = 10,000 elements large, of which 9,000 will be structural zeroes.
Applications of Historical MPMs
Why use historical MPMs, from a practical perspective? My own experiences with historical MPMs have shown me that they can lead to very different inferences than ahistorical MPMs. As an example, adaptive dynamics models of the evolution of sprouting frequency in two different species lead to dramatically different inferences when conducted using hMPMs than with ahMPMs. One of these species is Cypripedium parviflorum, the small yellow lady’s slipper, an endangered species occurring in the north-central portions of North America. This species is highly prone to become vegetatively dormant, during which it lacks any aboveground photosynthetic structures for potentially many years, and we had a 20yr census dataset for this analysis.
In that case, the ahMPM models suggested that dormancy made no evolutionary sense – sprouting in every year was the only adaptive solution. However, hMPMs led to models that showed that imperfect sprouting frequencies matching those observed in the wild were the most adaptive, representing an evolutionary stable strategy (ESS). The reason is that hMPMs allowed our analyses to incorporate life history relationships that ahMPMs did not (most notably costs of growth to survival and sprouting), and the addition of history also likely influenced the impact of density on population dynamics. This might also explain why population viability analyses (PVAs), which typically use MPMs, so frequently yield low quality or incorrect results.
In 2018, I published a paper that delved into the common demography and life history patterns associated with vegetative dormancy. As a part of this, I created historical MPMs for many species, but discovered that the lack of a standardized methodology for hMPM creation made the process extremely slow and often quite frustrating. This led me to start the project of developing this R package, and the result is a methodology that can yield a full set of historical matrices for any dataset within no more than 10-20 minutes from scratch. Learning materials are publicly available, and more are being produced. For example, instructional videos are available via the Shefferson lab website.
Package lefko3 is actively maintained and updated, and available on CRAN. Since release 3.0.0, which is described in this paper, we have added:
- Function-based age x stage matrices
- Environmental and individual covariate handling
- Re-parameterized negative binomial distribution, zero-inflated Poisson and negative binomial distributions as options for size and fecundity
- Function to test the assumptions of the Poisson distribution for size and fecundity, and expanded vignettes.
We are planning on adding stochastic analyses, density-dependent analyses, adaptive dynamics simulators, graphic error-checking methods, and alternate hMPM parameterizations. We encourage those wishing to explore complex matrix projection models to try this package out, particularly to test the impacts of individual history.
To read this application paper, check out the Methods in Ecology and Evolution article, ‘lefko3: Analysing individual history through size-classified matrix population models’.

