Our August Issue is now online! This issue contains 15 articles about the latest methods in ecology and evolution, including methods for estimating forest above-ground biomass, tracing essential amino acid assimilation, autonomous biodiversity surveys and much more! Read on to find out about this month’s featured articles and the article behind our wonderful whale cover.
Featured Articles
Estimating forest above-ground biomass with terrestrial laser scanning Improving the global monitoring of above-ground biomass (AGB) is crucial for forest management to be effective in climate mitigation. In the last decade, methods have been developed for estimating AGB from terrestrial laser scanning (TLS) data. TLS-derived AGB estimates can address current uncertainties in allometric and Earth observation (EO) methods that quantify AGB. In this article, Demol et al. assembled a global dataset of TLS scanned and consecutively destructively measured trees from a variety of forest conditions and reconstruction pipelines. They found that TLS-derived AGB closely agreed with destructive values, but identified below-average performances for smaller trees and conifers. In every individual study, TLS estimates of AGB were less biased and more accurate than those from allometric scaling models, especially for larger trees.
Deep learning as a tool for ecology and evolution Deep learning is gaining popularity in biology, where it has been used for automated species identification, environmental monitoring, ecological modelling, behavioural studies, DNA sequencing and population genetics and phylogenetics, among other applications. In this review, Borowiec et al. synthesise 818 studies using deep learning in the context of ecology and evolution to give a discipline-wide perspective necessary to promote a rethinking of inference approaches in the field.
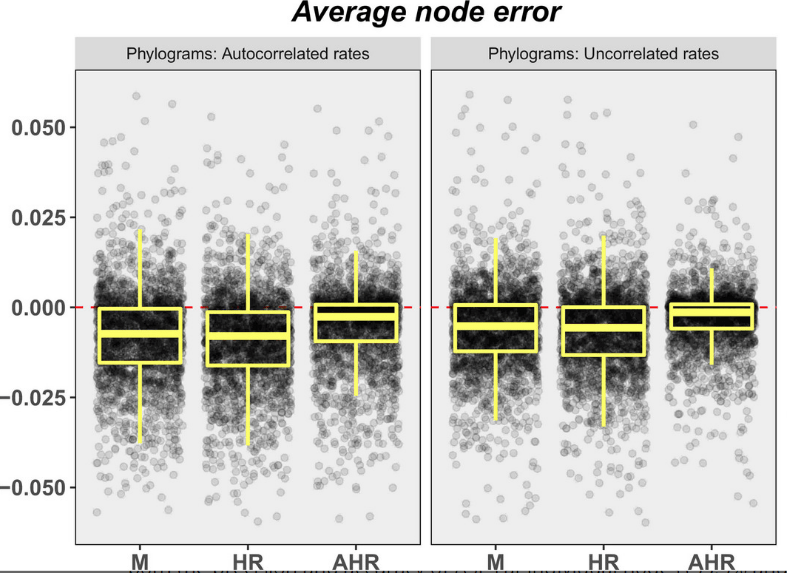
Chronogram or phylogram for ancestral state estimation? Modern methods of ancestral state estimation (ASE) incorporate branch length information, and it has been demonstrated that ASEs are more accurate when conducted on the branch lengths most correlated with a character’s evolution; however, a reliable method for choosing between alternate branch length sets for discrete characters has not yet been proposed. In this study, Wilson et al. simulate paired chronograms and phylograms, and generate binary characters that evolve in correlation with one of these. They then investigate (a) the effect of alternate branch lengths on ASE error and (b) whether phylogenetic signal statistics and/or model-fit statistic can be used to select the branch lengths most correlated with a binary character.
A continuous-score occupancy model Occupancy models are now widely used to account for false absences in field surveys and to reduce bias in estimates of covariate relationships. Existing occupancy models take as inputs binary detection/non-detection observations of species at each visit to each site. However, autonomous sensing devices and machine learning models are increasingly used to survey biodiversity, generating a new type of observation record that reflects the model’s confidence a species is present in each autonomously sensed file, instead of binary detection/non-detection data. These data are not directly compatible with traditional binary occupancy modelling methods. Here, Rhinehart et al. develop a new occupancy model that models continuous scores on a visit level as a Gaussian mixture, combining a distribution of scores for files that do contain the species of interest and a distribution of scores for files that do not.
Tracing essential amino acid assimilation and multichannel feeding Stable isotope ‘fingerprints’ are unique patterns in essential amino acid (EAA) δ13C values that vary consistently between energy channels like primary production and detritus, and they have emerged as a tool to trace energy flow in wild systems. Because animals cannot synthesize EAAs de novo and must acquire them from dietary proteins, ecologists often assume δ13C fingerprints travel through food webs unaltered. Numerous studies have used this approach to quantify energy flow and multichannel feeding in animals, but δ13C fingerprinting has never been experimentally tested in a vertebrate consumer. Here, Manlick & Newsome tested the efficacy of δ13C fingerprinting using captive deer mice Peromyscus maniculatus raised on diets containing bacterial, fungal and plant protein, as well as a combination of all three sources.
The Sperm Whale on the Cover
This month’s cover features a sperm whale in the deep azure waters of the Indian Ocean. There are many regions of the open ocean that have sparse survey coverage and for some species, such as sperm whales, there are few data on their distribution and habitat preferences. This lack of data hinders our ability to designate marine protected areas and manage the multiple threats from human activities. To address this knowledge gap, passive acoustic surveys on platforms of opportunity are key in generating data on cetaceans in remote regions. These surveys yield large amounts of data, the analysis of which is time consuming and can account for a substantial proportion of the survey budget. Semi-automatic processes enable the bulk of processing to be conducted automatically while allowing analyst time to be reserved for validating and correcting detections and classifications. In this issue, Webber et al. provide a streamlined method for analysing acoustic datasets so that such large-scale surveys can be more affordable, and therefore more accessible in future. Image credit: ©Greenpeace / Maarten van Rouveroy.